
This page uses JMOL Java technology to display interactive 3D models.
Most computers have Java already installed. If you do not have it, you can find it here: www.java.com
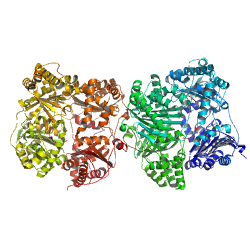 PDB id: 2G47, Human Insulin-Degrading Enzyme (IDE) PDB id: 2G47, Human Insulin-Degrading Enzyme (IDE)reference: Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. related entries: 2G48, 2G49, 2G54, 2G56, 2JBU, 2JG4 |
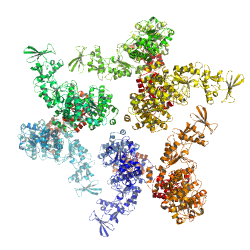 PDB id: 1XFU, Anthrax edema factor-Calmodulin complex
PDB id: 1XFU, Anthrax edema factor-Calmodulin complexreference: Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. related entries: 1XFV, 1XFW, 1XFX, 1XFY, 1XFZ, 1Y0V |
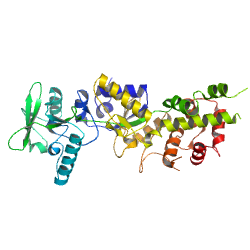 PDB id: 1YRT, Adenylyl cyclase toxin of Bordetella pertussis in complex with calmodulin PDB id: 1YRT, Adenylyl cyclase toxin of Bordetella pertussis in complex with calmodulinreference: Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin. related entries: 1YRU, 1ZOT, 2COL |
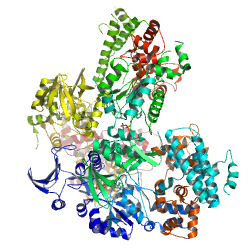 PDB id: 1PK0, Anthrax edema factor-calmodulin complexed with PMEApp PDB id: 1PK0, Anthrax edema factor-calmodulin complexed with PMEAppreference: Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection. |
 PDB id: 1SK6, Anthrax edema factor in complex with calmodulin, 3',5' cyclic AMP (cAMP), and pyrophosphate PDB id: 1SK6, Anthrax edema factor in complex with calmodulin, 3',5' cyclic AMP (cAMP), and pyrophosphatereference: Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate. |
 PDB id: 1K8T, Anthrax edema factor (EF)
PDB id: 1K8T, Anthrax edema factor (EF)
reference: Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin related entries: 1K90, 1K93, 1LVC |